The new edition of our “Analytical Sciences, Synthesis & Purification” catalog is available now! Offering a full suite of innovative products and technologies from Advion Interchim Scientific, this catalog features over 20,000 products ranging from consumables and reagents to instrumentation.
Not only a product resource, the catalog features in-depth scientific information to improve your daily workflow, including sample preparation tips and tricks, analysis technologies such as GC and LC/MS, and help selecting the best consumables for your workflow or purification.
Take advantage of this extensive guide to boost your lab with:
- 20,000 Products
- 12 Chapters
- Technicals Tips
- New Systems
Authors: University of Birmingham, United Kingdom
Abstract
The ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter cloacae) represent clinically important bacterial species that are responsible for most hospital-acquired drug-resistant infections; hence, the need for rapid identification is of high importance. Previous work has demonstrated the suitability of liquid extraction surface analysis mass spectrometry (LESA-MS) for the direct analysis of colonies of two of the ESKAPE pathogens (Staphylococcus aureus and Pseudomonas aeruginosa) growing on agar. Here, we apply LESA-MS to the remaining four ESKAPE species (E. faecium E745, K. pneumoniae KP257, A. baumannii AYE, and E. cloacae S11) as well as E. faecalis V583 (a close relative of E. faecium) and a clinical isolate of A. baumannii AC02 using an optimized solvent sampling system. In each case, top-down LESA MS/MS was employed for protein identification. In total, 24 proteins were identified from 37 MS/MS spectra by searching against protein databases for the individual species. The MS/MS spectra for the identified proteins were subsequently searched against multiple databases from multiple species in an automated data analysis workflow with a view to determining the accuracy of identification of unknowns. Out of 24 proteins, 19 were correctly assigned at the protein and species level, corresponding to an identification success rate of 79%.
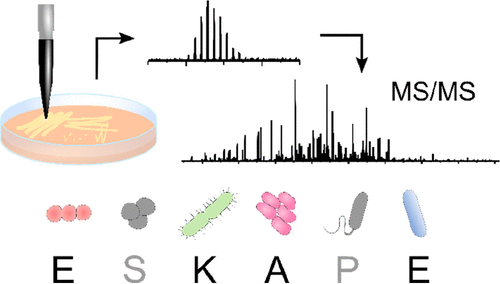
LESA-MS was performed using the Advion Interchim Scientific® TriVersa NanoMate®.
Western Washington University
Abstract
Sortase‐catalyzed transacylation reactions are widely used for the construction of non‐natural protein derivatives. However, the most commonly used enzyme for these strategies (sortase A from Staphylococcus aureus) is limited by its narrow substrate scope. To expand the range of substrates compatible with sortase‐mediated reactions, we characterized the in vitro substrate preferences of eight sortase A homologues.
From these studies, we identified sortase A enzymes that recognize multiple substrates that are unreactive toward sortase A from S. aureus.
We further exploited the ability of sortase A from Streptococcus pneumoniae to recognize an LPATS substrate to perform a site‐specific modification of the N‐terminal serine residue in the naturally occurring antimicrobial peptide DCD‐1L.
Finally, we unexpectedly observed that certain substrates (LPATXG, X=Nle, Leu, Phe, Tyr) were susceptible to transacylation at alternative sites within the substrate motif, and sortase A from S. pneumoniae was capable of forming oligomers. Overall, this work provides a foundation for the further development of sortase enzymes for use in protein modification.
Mass spectrometry analysis was performed using the Advion Interchim Scientific® expression® Compact Mass Spectrometer (CMS).
Nirma University, India; Rega Institute for Medical Research
Abstract
In order to design novel anti-HIV agents, pharmacophore modelling, virtual screening, 3D-QSAR and molecular docking studies were performed. 50 quinoxaline derivatives with different substitutions were designed on basis of both ligand based drug design approaches and were mapped on the best pharmacophore model. From this, best 32 quinoxaline derivatives were docked onto the active site of integrase enzyme and in-silico ADMET properties were also predicted. From this data, synthesis of top 7 quinoxaline derivatives was carried out and were characterized using Mass, 1H-NMR and 13C-NMR spectroscopy. Purity of compounds were checked using HPLC.
Two quinoxaline derivatives showed good results, which can be further explored to develop novel anti-HIV agents.
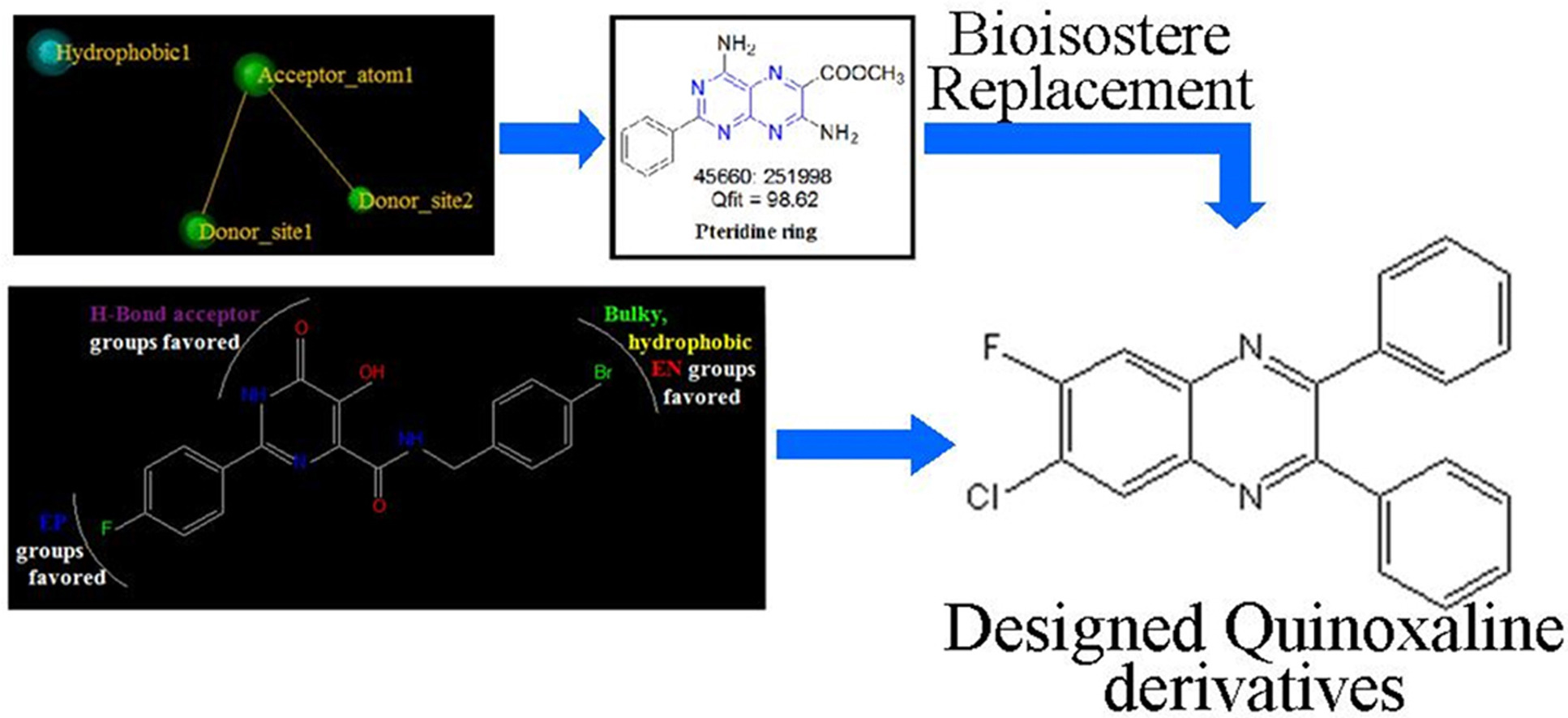
Lyon College, University of Rhode Island
Abstract
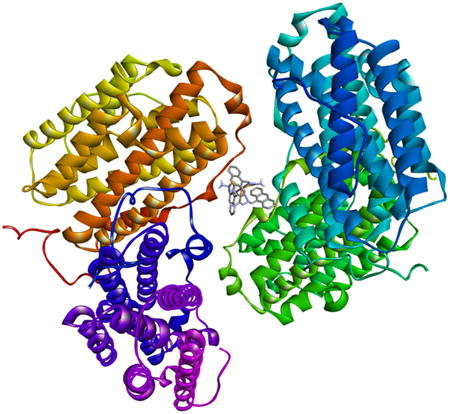
MOLSTR, Accepted manuscript. doi:10.1016/j.molstruc.2016.05.075
We have synthesized a trio of gallium complexes bearing 9-anthraldehyde thiosemicarbazones. The complexes were assessed for their anticancer activity and their biophysical reactivity was also investigated.
The results of docking studies with DNA, ribonucleotide reductase and human serum albumin, however showed that the complex with the best biological activity had the largest binding constant to DNA.
Electrospray ionization mass spectrometry (ESI-MS) was recorded using the Advion expression Compact Mass Spectrometer (CMS).
Xention Limited, United Kingdom; University of Ljubljana, Slovenia
Abstract
The rodent neuroblastoma cell line, ND7-23, is used to express voltage-dependent sodium (Nav) and other neuronal ion channels resistant to heterologous expression in Chinese hamster ovary (CHO) or human embryonic kidney (HEK) cells. Their advantage is that they provide endogenous factors and signaling pathways to promote ion channel peptide folding, expression, and function at the cell surface and are also amenable to automated patch clamping. However, ND7-23 cells exhibit endogenous tetrodotoxin (TTX)-sensitive Nav currents, and molecular profiling has revealed the presence of Nav1.2, Nav1.3, Nav1.6, and Nav1.7 transcripts, but no study has determined which subtypes contribute to functional channels at the cell surface. In this publication, we profiled the repertoire of functional Nav channels endogenously expressed in ND7-23 cells using the QPatch automated patch clamp platform and selective toxins and small molecules.
Mass spec analysis was performed using the Advion expression Compact Mass Spectrometer (CMS).
University of Surrey, Surrey and Borders Partnership NHS Foundation Trust, Advion, Ltd.
Abstract
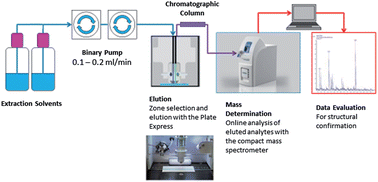
Surface mass spectrometry methods can be difficult to use effectively with low cost, portable mass spectrometers. A new analytical method is presented where analytes are extracted from a sample using a solvent flushed across the surface under high pressure, separated using a chromatography column and then analysed using the Advion Interchim Scientific® expression® Compact Mass Spectrometer (CMS). In this publication, we demonstrate the efficacy of the method for the quantitative detection of cocaine and benzoylecgonine in urine and oral fluid.
In this note, a quick and direct screening method for the detection of illicit drugs on paper currency is introduced using the Touch Express™ Open Port Sampling Interface (OPSI) on the Advion expression® Compact Mass Spectrometer (CMS). A range of paper currency bills from different countries were analyzed for traces of illicit drugs.

Authors: Heinrich Pette Institute – Leibniz Institute for Experimental Virology, European XFEL GmbH, Vienna University of Technology (TU Wien), Indiana University, University Medical Center Hamburg-Eppendorf, Bernard Nocht Institute for Tropical Medicine and German Center for Infection Research
Abstract
Noroviruses cause immense sporadic gastroenteritis outbreaks worldwide. Emerging genotypes, which are divided based on the sequence of the major capsid protein VP1, further enhance this public threat. Self-assembling properties of the human norovirus major capsid protein VP1 are crucial for using virus-like particles (VLPs) for vaccine development. However, there is no vaccine available yet. Here, VLPs from different variants produced in insect cells were characterized in detail using a set of biophysical and structural tools. We used native mass spectrometry, gas-phase electrophoretic mobility molecular analysis, and proteomics to get clear insights into particle size, structure, and composition, as well as stability. Generally, noroviruses have been known to form mainly T = 3 particles. Importantly, we identified a major truncation in the capsid proteins as a likely cause for the formation of T = 1 particles. For vaccine development, particle production needs to be a reproducible, reliable process. Understanding the underlying processes in capsid size variation will help to produce particles of a defined capsid size presenting antigens consistent with intact virions. Next to vaccine production itself, this would be immensely beneficial for bio-/nano-technological approaches using viral particles as carriers or triggers for immunological reactions.
This publication features the Advion TriVersa NanoMate® as an automated nano-ESI source for CDMS measurements.

University of Birmingham
Abstract
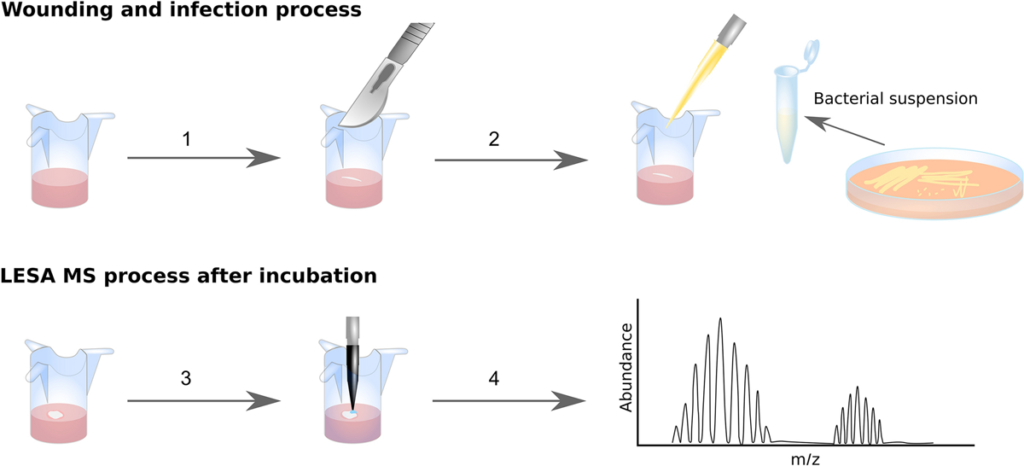 Figure 1.
Figure 1.
Trauma is one of the leading causes of death in people under the age of 49 and complications due to wound infection are the primary cause of death in the first few days after injury. The ESKAPE pathogens are a group of bacteria that are a leading cause of hospital-acquired infections and a major concern in terms of antibiotic resistance. Here, we demonstrate a novel and highly accurate approach for the rapid identification of ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) directly from infected wounds in 3D in vitro skin models. Wounded skin models were inoculated with bacteria and left to incubate. Bacterial proteins were identified within minutes, directly from the wound, by liquid extraction surface analysis mass spectrometry. This approach was able to distinguish closely related strains and, unlike genomic approaches, can be modified to provide dynamic information about pathogen behaviour at the wound site. In addition, since human skin proteins were also identified, this method offers the opportunity to analyse both host and pathogen biomarkers during wound infection in near real-time.







