The brain is a remarkably complex organ and cholesterol homeostasis underpins brain function. It is known that cholesterol is not evenly distributed across different brain regions; however, the precise map of cholesterol metabolism in the brain remains unclear. If cholesterol metabolism is to be correlated with brain function it is essential to generate such a map. Here we describe an advanced mass spectrometry platform to reveal spatial cholesterol metabolism in situ at 400-µm spot diameter on 10-µm tissue slices from mouse brain. We mapped, not only cholesterol, but also other biologically active sterols arising from cholesterol turnover in both wild type and mice lacking cholesterol 24S-hydroxylase (CYP46A1), the major cholesterol metabolizing enzyme.
Fully automated chip-based nanoelectrospray ionization-mass spectrometry as an effective tool for rapid and high-throughput screening of 5α-reductase inhibitors
Shanghai University of Traditional Chinese Medicine
Abstract
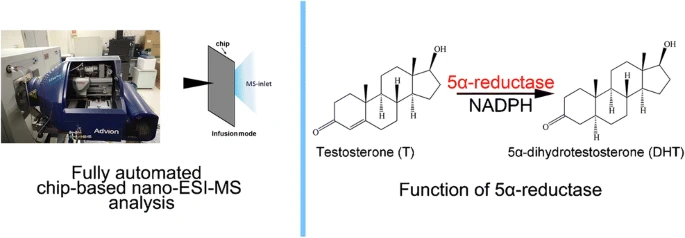
The 5α-reductase converts testosterone to dihydrotestosterone (DHT), and excess DHT could cause androgen-related diseases such as androgenetic alopecia and benign prostatic hyperplasia (BPH). To discover new 5α-reductase inhibitors, effective drug screening method with high throughput is thus essential. In this study, fully automated chip-based nanoelectrospray ionization-mass spectrometry (nano-ESI-MS) was innovatively utilized as a screening tool for 5α-reductase inhibitory assay in direct infusion mode, which simplified sample pretreatment and greatly improved experimental efficiency. The preliminary data indicated that curcumin, a natural anti-inflammatory compound, exhibited notably 5α-reductase inhibition activity. Moreover, the obtained results of the chip-based nano-ESI-MS were well consistent with those of HPLC-MS, which suggested that the chip-based nano-ESI-MS could be treated as a rapid and high-throughput drugs screening strategy in pharmaceutical development.
Total Fatty Acid Analysis of Human Blood Samples in One Minute by High-Resolution Mass Spectrometry
Easy, Fast, and Reproducible Quantification of Cholesterol and Other Lipids in Human Plasma by Combined High Resolution MSX and FTMS Analysis
Oxidative modification of skin lipids by cold atmospheric plasma (CAP): A standardizable approach using RP-LC/MS2 and DI-ESI/MS2 – Using the TriVersa NanoMate
The Advion TriVersa NanoMate nanoelectrospray ionization technology was selected as the ion source for its long, stable spray capabilities ideal for lipid analysis.
Abstract
Cold atmospheric plasma (CAP) is an emerging source for the locally defined delivery of reactive species, and its clinical potential has been identified in the control of inflammatory processes, such as acute and chronic wounds, or cancerous lesions. Lipids, due to their localization and chemical structure as ideal targets for oxidative species, are relevant modifiers of physiological processes. Human forehead lipids collected on a target were treated by an argon plasma jet and immediately analyzed by direct-infusion high-resolution tandem mass spectrometry (DI-MS2) or liquid chromatography-tandem MS (RP-LC/MS2). Subsequent data analysis was performed by LipidHunter (University of Leipzig), LipidXplorer (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden), and LipidSearch (TS). With either MS method, all major lipid classes of sebum lipids were detected. Significant differences regarding triacylglycerols (predominantly identified in RP-LC/MS2) and ceramides (predominantly identified in DI-MS2) indicate experimental- or approach-inherent distinctions. A CAP-driven oxidation of triacyclglycerols, ceramides, and cholesteryl esters was detected such as truncations and hydroperoxylations, but at a significantly lower extent than expected. Scavenging of reactive species due to naturally present antioxidants in the samples and the absence of a liquid interphase to allow reactive species deposition by the CAP will have contributed to the limited amount of oxidation products observed. In addition, limitations of the software’s capability of identifying unexpected oxidized lipids potentially led to an underestimation of the CAP impact on skin lipids, indicating a need for further software development. With respect to the clinical application of CAP, the result indicates that intact skin with its sebum/epidermal lipid overlay is well protected and that moderate treatment will yield limited (if any) functional consequences in the dermal tissue.
University of Cambridge, NIHR BCR Metabolomics and Lipidomics Facility
What is the focus of your lab’s research?
Almost exclusively high throughput lipid profiling of samples from large epidemiological studies, where we study gene-lifestyle interactions. The method works with plasma samples as well as dried blood spots. The method is also applied to small-scale studies of specific disease groups, dietary interventions and model systems such as yeast. The TriVersa NanoMate® is also used to study the lipid composition of tissues analyzed by LESA®.
Why did you incorporate the TriVersa NanoMate® into your laboratory?
The TriVersa NanoMate® is essential to efficiently analyze large-scale studies. It offers a really robust method for high throughput studies with minimal carryover.
Who would you recommend to purchase the TriVersa NanoMate®?
I recommend the TriVersa NanoMate® to laboratories with a large number of samples requiring a robust and reliable delivery system. The TriVersa NanoMate® eliminates typical nanoelectrospray ionization challenges.
Do you have any publications or presentations using the TriVersa NanoMate®?
Publication Highlight:
Development and Application of High-Throughput Single Cell Lipid Profiling: A Study of SNCA-A53T Human Dopamine Neurons
Snowden, et al. iScience, 2020, 23(10), 101703
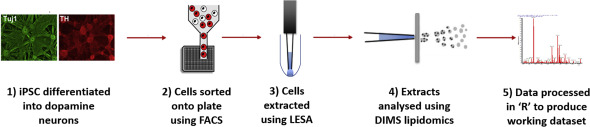
Combining FACS and LESA-MS to establish high-throughput single cell lipid profiling. Research identifies lipid differences found within and between populations of human dopamine neurons.
Other Publications:
- Snowden et al. Combining lipidomics and machine learning to measure clinical lipids in dried blood spots. Metabolomics. DOI: 10.1007/s11306-020-01703-0
- Koulman et al. The development and validation of a fast and robust dried blood spot based lipid profiling method to study infant metabolism. Metabolomics. DOI: 10.1007/s11306-014-0628-z
- Furse et al. A high-throughput platform for detailed lipidomic analysis of a range of mouse and human tissues. Analytical and Bioanalytical Chemistry. DOI: 10.1007/s00216-020-02511-0
- Harshfield et al. An unbiased lipid phenotyping approach to study the genetic determinants of lipids and their associations with coronary heart disease risk factors. J Proteom Res. DOI: 10.1021/acs.jproteome.8b00786
- Mann et al. Insights into genetic variants associated with NASH-fibrosis from metabolite profiling. Hum Mol Genet. DOI: 10.1093/hmg/ddaa162
Minimally-destructive atmospheric ionisation mass spectrometry authenticates authorship of historical manuscripts
University of Glasgow, University of Edinburgh
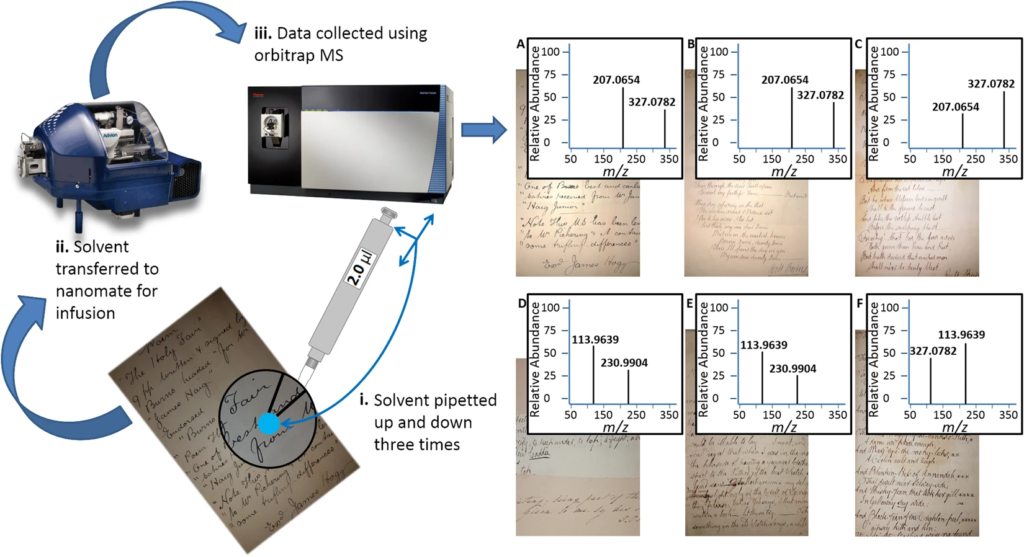
Authentic historic manuscripts fetch high sums, but establishing their authenticity is challenging, relies on a host of stylistic clues and requires expert knowledge. High resolution mass spectrometry has not, until now, been applied to guide the authentication of historic manuscripts. Robert Burns is a well-known Scottish poet, whose fame, and the eponymous ‘Burns Night’ are celebrated world-wide. Authenticity of his works is complicated by the ‘industrial’ production of fakes by Alexander Smith in the 1890s, many of which were of good quality and capable of fooling experts. This study represents the first analysis of the inks and paper used in Burns poetry, in a minimally destructive manner that could find application in many areas. Applying direct infusion mass spectrometry to a panel of selected authenticated Burns and Smith manuscripts, we have produced a Support Vector Machine classifier that distinguishes Burns from Smith with a 0.77 AUC. Using contemporary recipes for inks, we were also able to match features of each to the inks used to produce some of Burns’ original manuscripts. We anticipate the method and classifier having broad application in authentication of manuscripts, and our analysis of contemporary inks to provide insights into the production of written works of art.
Liquid Extraction Surface Analysis Mass Spectrometry (LESA-MS): Examples of a New Surface Probing Technique for Clinical and Pre-Clinical Applications
Executive Summary:
- LESA and LESAplus liquid chromatography provide novel surface analysis tools for the spatial resolution of analytes from a wide variety of surfaces.
- Whole body or organ sections in particular can be probed for small molecules, lipids and proteins with a resolution of 400 – 1000μm.
- Commercial solution allows for automated processing of samples (Advion TriVersa NanoMate® LESA®).
This poster was presented at MSACL 2018 US in Palm Springs, CA.
Protein Identification in Imaging Mass Spectrometry through Spatially Targeted Liquid Micro-Extractions featuring the Advion TriVersa NanoMate® LESA® Plus
Daniel J. Ryan, David Nei, Boone M. Prentice, Kristie L. Rose, Richard M. Caprioli, Jeffrey M. Spraggins
Robotic liquid surface extractions can be used to interrogate discrete regions of tissue to provide protein identifications with high throughput, accuracy, and robustness. The direct coupling of tissue surface extractions and liquid chromatography, offers a new and effective approach to provide spatial proteomics data in an imaging experiment.
Tissue extractions were completed using the TriVersa NanoMate® (Advion, Inc., Ithaca, NY, USA) modified to include a glass capillary (LESA Plus) for improved spatial resolution and online integration with LC-based experiments. 36 Scanned images of thaw-mounted samples were uploaded to the ChipSoft Software (Advion, Inc.) to allow histological regions of interest to be selected for analysis.
Integration of Ion Mobility MSE after Fully Automated, Online, High- Resolution Liquid Extraction Surface Analysis Micro-Liquid Chromatography featuring the Advion TriVersa NanoMate® LESA® Plus
Lieke Lamont, Mark Baumert, Nina Ogrinc Potočnik, Mark Allen, Rob Vreefken, Ron M. A. Heeren, and Tiffany Porta
Direct analysis by mass spectrometry (imaging) has become increasingly deployed in preclinical and clinical research due to its rapid and accurate readouts. However, when it comes to biomarker discovery or histopathological diagnostics, more sensitive and in-depth profi ling from localized areas is required. We developed a comprehensive, fully automated online platform for high-resolution liquid extraction surface analysis (HR-LESA) followed by micro−liquid chromatography (LC) separation and a data-independent acquisition strategy for untargeted and low abundant analyte identifi cation directly from tissue sections. Applied to tissue sections of rat pituitary, the platform demonstrated improved spatial resolution, allowing sample areas as small as 400 μ m to be studied, a major advantage over conventional LESA.
The LESA extraction was performed using the automated TriVersa NanoMate® The LESA extraction was controlled by a beta version of the LESA Plus software (Advion, UK)



